A list of publications from my lab and with my collaborators.
2022

Ivanov, Alexander; Willett, Wesley; Jacob, Christian
EvoIsland: interactive evolution via an island-inspired spatial user interface framework Proceedings Article
In: Proceedings of the Genetic and Evolutionary Computation Conference, pp. 1200–1208, 2022.
@inproceedings{ivanov2022evoisland,
title = {EvoIsland: interactive evolution via an island-inspired spatial user interface framework},
author = {Alexander Ivanov and Wesley Willett and Christian Jacob},
doi = {https://doi.org/10.1145/3512290.3528722},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
booktitle = {Proceedings of the Genetic and Evolutionary Computation Conference},
pages = {1200–1208},
abstract = {We present EvoIsland, a scalable interactive evolutionary user interface framework inspired by the spatially isolated land masses seen on Earth. Our generalizable interaction system encourages creators to spatially explore a wide range of design possibilities through the combination, separation, and rearrangement of hexagonal tiles on a grid. As these tiles are grouped into islandlike clusters, localized populations of designs form through an underlying evolutionary system. The interactions that take place within EvoIsland provide content creators with new ways to shape, display and assess populations in evolutionary systems that produce a wide range of solutions with visual phenotype outputs.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
2020
Brintnell, Erin; Brierley, Owen; Christensen, Neil; Jacob, Christian
Balancing simulation and gameplay — applying game user research to LeukemiaSIM Technical Report
2020.
@techreport{nokey,
title = {Balancing simulation and gameplay — applying game user research to LeukemiaSIM},
author = {Erin Brintnell and Owen Brierley and Neil Christensen and Christian Jacob},
url = {http://arxiv.org/abs/2009.11404},
year = {2020},
date = {2020-09-23},
urldate = {2020-09-23},
abstract = {A bioinformatics researcher and a game design researcher walk into a lab... This paper shares two case-studies of a collaboration between a bioinformatics researcher who is developing a set of educational VR simulations for youth and a consultative game design researcher with a background in Games User Research (GUR) techniques who assesses and iteratively improves the player experience in the simulations. By introducing games-based player engagement strategies, the two researchers improve the (re)playability of these VR simulations to encourage greater player engagement and retention.},
keywords = {},
pubstate = {published},
tppubtype = {techreport}
}
2019

Eidelberg, Eric; Jacob, Christian; Denzinger, Jorg
Using Active Probing by a Game Management AI to Faster Classify Players Conference
2019 IEEE Conference on Games (CoG), IEEE, London, UK, 2019, ISBN: 978-1-72811-884-0.
@conference{nokey,
title = {Using Active Probing by a Game Management AI to Faster Classify Players},
author = {Eric Eidelberg and Christian Jacob and Jorg Denzinger},
url = {https://ieeexplore.ieee.org/document/8847957/},
doi = {10.1109/CIG.2019.8847957},
isbn = {978-1-72811-884-0},
year = {2019},
date = {2019-08-01},
urldate = {2019-08-01},
booktitle = {2019 IEEE Conference on Games (CoG)},
pages = {1-8},
publisher = {IEEE},
address = {London, UK},
abstract = {In this paper, we present the use of a so-called Game Management AI to classify players not just by passively observing them, but by actively manipulating the game to get the players to provide data currently missing to achieve the classification. We call this “Active Probing”. The Game Management AI uses two sets of rules, one set that contains rules that are intended to represent the knowledge allowing a classification and one set that contains rules that indicate which game events can contribute to triggering conditions used in the first rule set. When a rule of the first set comes near to being triggered, the event suggested by an appropriate rule in the second set is then offered to the player in the game.},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}

Ivanov, Alexander; Danyluk, Kurtis; Jacob, Christian; Willett, Wesley
A Walk Among the Data: Exploration and Anthropomorphism in Immersive Unit Visualizations Journal Article
In: IEEE Computer Graphics and Applications, vol. Special Issue May/June 2019, 2019.
@article{Ivanov2019,
title = {A Walk Among the Data: Exploration and Anthropomorphism in Immersive Unit Visualizations},
author = {Alexander Ivanov and Kurtis Danyluk and Christian Jacob and Wesley Willett},
doi = {10.1109/MCG.2019.2898941},
year = {2019},
date = {2019-05-01},
urldate = {2019-05-01},
journal = {IEEE Computer Graphics and Applications},
volume = {Special Issue May/June 2019},
abstract = {We examine the potential for immersive unit visualizations – interactive virtual environments populated with objects representing individual items in a dataset. Our explorations highlight how immersive unit visualizations in virtual reality can allow viewers to examine data at multiple scales,
support immersive exploration, and create affective, personal experiences with data.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
support immersive exploration, and create affective, personal experiences with data.

Kobsar, D.; Osis, S. T.; Jacob, Christian; Ferber, Reed
Validity of a novel method to measure vertical oscillation during running using a depth camera Journal Article
In: Journal of Biomechanics, vol. 85, pp. 182-186, 2019.
@article{Kobsar2019,
title = {Validity of a novel method to measure vertical oscillation during running using a depth camera},
author = {D. Kobsar and S.T. Osis and Christian Jacob and Reed Ferber },
doi = {10.1016/j.jbiomech.2019.01.006},
year = {2019},
date = {2019-03-06},
urldate = {2019-03-06},
journal = {Journal of Biomechanics},
volume = {85},
pages = {182-186},
abstract = {Recent advancements in low-cost depth cameras may provide a clinically accessible alternative to conventional three-dimensional (3D) multi-camera motion capture systems for gait analysis. However, there remains a lack of information on the validity of clinically relevant running gait parameters such as vertical oscillation (VO). The purpose of this study was to assess the validity of measures of VO during running gait using raw depth data, in comparison to a 3D multi-camera motion capture system. Sixteen healthy adults ran on a treadmill at a standard speed of 2.7 m/s. The VO of their running gait was simultaneously collected from raw depth data (Microsoft Kinect v2) and 3D marker data (Vicon multi-camera motion capture system). The agreement between the VO measures obtained from the two systems was assessed using a Bland-Altman plot with 95% limits of agreement (LOA), a Pearson’s correlation coefficient (r), and a Lin’s concordance correlation coefficient (rc). The depth data from the Kinect v2 demonstrated excellent results across all measures of validity (r = 0.97; rc = 0.97; 95% LOA = −8.0 mm – 8.7 mm), with an average absolute error and percent error of 3.7 (2.1) mm and 4.0 (2.0)%, respectively. The findings of this study have demonstrated the ability of a low cost depth camera and a novel tracking method to accurately measure VO in running gait.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Davison, Timothy; Samavati, Faramarz; Jacob, Christian
Lifebrush: painting, simulating, and visualizing dense biomolecular environments Journal Article
In: Computers & Graphics, vol. 82, pp. 232–242, 2019.
@article{davison2019lifebrush,
title = {Lifebrush: painting, simulating, and visualizing dense biomolecular environments},
author = {Timothy Davison and Faramarz Samavati and Christian Jacob},
doi = {https://doi.org/10.1016/j.cag.2019.05.006},
year = {2019},
date = {2019-01-01},
urldate = {2019-01-01},
journal = {Computers & Graphics},
volume = {82},
pages = {232--242},
publisher = {Elsevier},
abstract = {LifeBrush is a Cyberworld for painting dynamic molecular illustrations in virtual reality (VR) that then come to life as interactive simulations. We designed our system for the biological mesoscale, a spatial scale where molecules inside cells interact to form larger structures and execute the functions of cellular life. We bring our immersive illustrations to life in VR using agent-based modelling and simulation. Our sketch-based brushes use discrete element texture synthesis to generate molecular-agents along the brush path derived from examples in a palette. In this article we add a new tool to sculpt the geometry of the environment and the molecules. We also introduce a new history based visualization that enables the user to interactively explore and distil, from the busy and chaotic mesoscale environment, the interactions between molecules that drive cellular processes. We demonstrate our system with a mitochondrion example.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2018

Davison, Timothy; Samavati, Faramarz; Jacob, Christian
LifeBrush: Painting Interactive Agent-based Simulations Conference
2018 International Conference on Cyberworlds (CW), IEEE 2018.
@conference{Davison2018,
title = {LifeBrush: Painting Interactive Agent-based Simulations},
author = {Timothy Davison and Faramarz Samavati and Christian Jacob},
year = {2018},
date = {2018-10-03},
urldate = {2018-10-03},
booktitle = {2018 International Conference on Cyberworlds (CW)},
organization = {IEEE},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}

Wrobleski, Brad; Ivanov, Alexander; Eidelberg, Eric; Etemad, Katayoon; Gadbois, Denis; Jacob, Christian
Lucida: Enhancing the Creation of Photography through Semantic, Sympathetic, Augmented Voice Agent Interaction Proceedings Article
In: International Conference on Human Computer Interaction, pp. 200-216, Springer 2018.
@inproceedings{B2018,
title = {Lucida: Enhancing the Creation of Photography through Semantic, Sympathetic, Augmented Voice Agent Interaction},
author = {Brad Wrobleski and Alexander Ivanov and Eric Eidelberg and Katayoon Etemad and Denis Gadbois and Christian Jacob },
doi = {10.1007/978-3-319-91250-9_16},
year = {2018},
date = {2018-07-15},
urldate = {2018-07-15},
booktitle = {International Conference on Human Computer Interaction},
pages = {200-216},
organization = {Springer},
abstract = {Abstract. We present a dynamic framework for the integration of Machine
Learning (ML), Augmented Reality (AR), Affective Computing
(AC), Natural Language Processing (NLP) and Computer Vision (CV)
to make possible, the development of a mobile, sympathetic, ambient
(virtual), augmented intelligence (Agent). For this study we developed a
prototype agent to assist photographers to enhance the learning and
creation of photography. Learning the art of photography is complicated
by the technical complexity of the camera, the limitations of the user to
see photographically and the lack of real time instruction and emotive
support. The study looked at the interaction patterns between human
student and instructor, the disparity between human vision and the camera,
and the potential of an ambient agent to assist students in learning.
The study measured the efficacy of the agent and its ability to transmute
human-to-Human method of instruction to human-to-Agent interaction.
This study illuminates the effectiveness of Agent based instruction. We
demonstrate that a mobile, semantic, sympathetic, augmented intelligence,
ambient agent can ameliorate learning photography metering in
real time, ’on location’. We show that the integration of specific technologies
and design produces an effective architecture for the creation of
augmented agent-based instruction.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
Learning (ML), Augmented Reality (AR), Affective Computing
(AC), Natural Language Processing (NLP) and Computer Vision (CV)
to make possible, the development of a mobile, sympathetic, ambient
(virtual), augmented intelligence (Agent). For this study we developed a
prototype agent to assist photographers to enhance the learning and
creation of photography. Learning the art of photography is complicated
by the technical complexity of the camera, the limitations of the user to
see photographically and the lack of real time instruction and emotive
support. The study looked at the interaction patterns between human
student and instructor, the disparity between human vision and the camera,
and the potential of an ambient agent to assist students in learning.
The study measured the efficacy of the agent and its ability to transmute
human-to-Human method of instruction to human-to-Agent interaction.
This study illuminates the effectiveness of Agent based instruction. We
demonstrate that a mobile, semantic, sympathetic, augmented intelligence,
ambient agent can ameliorate learning photography metering in
real time, ’on location’. We show that the integration of specific technologies
and design produces an effective architecture for the creation of
augmented agent-based instruction.
Kelly, Justin; Jacob, Christian
evoExplore: Multiscale Visualization of Evolutionary Histories in Virtual Reality Proceedings Article
In: Evolutionary and Biologically Inspired Music, Sound, Art and Design, pp. 112-127, Springer Nature, 2018.
@inproceedings{evoStar2018,
title = {evoExplore: Multiscale Visualization of Evolutionary Histories in Virtual Reality},
author = {Justin Kelly and Christian Jacob},
doi = {https://doi.org/10.1007/978-3-319-77583-8_8},
year = {2018},
date = {2018-03-07},
urldate = {2018-03-07},
booktitle = {Evolutionary and Biologically Inspired Music, Sound, Art and Design},
pages = {112-127},
publisher = {Springer Nature},
abstract = {evoExplore is a system built for virtual reality (VR) and designed to assist evolutionary design projects. Built with the Unity 3D game engine and designed with future development and expansion in mind, evoExplore allows the user to review and visualize data collected from evolutionary design experiments. Expanding upon existing work, evoExplore provides the tools needed to breed your own evolving populations of designs, save the results from such evolutionary experiments and then visualize the recorded data as an interactive VR experience. evoExplore allows the user to dynamically explore their own evolutionary experiments, as well as those produced by other users. In this document we describe the features of evoExplore, its use of virtual reality and how it supports future development and expansion.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
2016

Yuen, Douglas; Cartwright, Stephen; Jacob, Christian
Eukaryo: Virtual Reality Simulation of a Cell Proceedings Article
In: Proceedings of the 2016 Virtual Reality International Conference, pp. 3:1–3:4, ACM, Laval, France, 2016, ISBN: 978-1-4503-4180-6.
@inproceedings{Yuen:2016:EVR:2927929.2927931,
title = {Eukaryo: Virtual Reality Simulation of a Cell},
author = {Douglas Yuen and Stephen Cartwright and Christian Jacob},
doi = {10.1145/2927929.2927931},
isbn = {978-1-4503-4180-6},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
booktitle = {Proceedings of the 2016 Virtual Reality International Conference},
pages = {3:1--3:4},
publisher = {ACM},
address = {Laval, France},
series = {VRIC '16},
abstract = {Eukaryo is an interactive, 3-dimensional, simulated bio-molecular world that allows users to explore the complex environment within a biological cell. Eukaryo was developed using Unity, leveraging the capabilities and high performance of a commercial game engine. Through the use of MiddleVR, our tool can support a wide variety of interaction platforms including 3D virtual reality (VR) environments, such as head-mounted displays and large scale immersive visualization facilities.
Our model demonstrates key structures of a generic eukaryotic cell. Users are able to use multiple modes to explore the cell, its structural elements, its organelles, and some key metabolic processes. In contrast to textbook diagrams and even videos, Eukaryo immerses users directly in the biological environment giving a more effective demonstration of how cellular processes work, how compartmentalization affects cellular functions, and how the machineries of life operate.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
Our model demonstrates key structures of a generic eukaryotic cell. Users are able to use multiple modes to explore the cell, its structural elements, its organelles, and some key metabolic processes. In contrast to textbook diagrams and even videos, Eukaryo immerses users directly in the biological environment giving a more effective demonstration of how cellular processes work, how compartmentalization affects cellular functions, and how the machineries of life operate.

Yuen, Douglas; Santoso, Markus; Cartwright, Stephen; Jacob, Christian
Eukaryo: An AR and VR Application for Cell Biology. Journal Article
In: International Journal of Virtual Reality, vol. 16, no. 1, 2016.
@article{yuen2016eukaryo,
title = {Eukaryo: An AR and VR Application for Cell Biology.},
author = {Douglas Yuen and Markus Santoso and Stephen Cartwright and Christian Jacob},
doi = {10.1145/2927929.2927931},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
journal = {International Journal of Virtual Reality},
volume = {16},
number = {1},
abstract = {Eukaryo is an interactive, 3-dimensional, simulated bio-molecular world that allows users to explore the complex environment within a biological cell. Eukaryo was developed using Unity, leveraging the capabilities and high performance of a commercial game engine. Through the use of MiddleVR, our tool can support a wide variety of interaction platforms including 3D virtual reality (VR) environments, such as head-mounted displays and large scale immersive visualization facilities.
Our model demonstrates key structures of a generic eukaryotic cell. Users are able to use multiple modes to explore the cell, its structural elements, its organelles, and some key metabolic processes. In contrast to textbook diagrams and even videos, Eukaryo immerses users directly in the biological environment giving a more effective demonstration of how cellular processes work, how compartmentalization affects cellular functions, and how the machineries of life operate.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Our model demonstrates key structures of a generic eukaryotic cell. Users are able to use multiple modes to explore the cell, its structural elements, its organelles, and some key metabolic processes. In contrast to textbook diagrams and even videos, Eukaryo immerses users directly in the biological environment giving a more effective demonstration of how cellular processes work, how compartmentalization affects cellular functions, and how the machineries of life operate.
Kelly, Justin; Jacob, Christian
evoVision3D: A Multiscale Visualization of Evolutionary Histories Proceedings Article
In: International Conference on Parallel Problem Solving from Nature, pp. 942–951, Springer 2016.
@inproceedings{kelly2016evovision3d,
title = {evoVision3D: A Multiscale Visualization of Evolutionary Histories},
author = {Justin Kelly and Christian Jacob},
doi = {10.1007/978-3-319-45823-6_88},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
booktitle = {International Conference on Parallel Problem Solving from Nature},
pages = {942--951},
organization = {Springer},
abstract = {Evolutionary computation is a field defined by large data sets and complex relationships. Because of this complexity it can be difficult to identify trends and patterns that can help improve future projects and drive experimentation. To address this we present evoVision3D, a multiscale 3D system designed to take data sets from evolutionary design experiments and visualize them in order to assist in their inspection and analysis. Our system is implemented in the Unity 3D game development environment, for which we show that it lends itself to immersive navigation through large data sets, going even beyond evolution-based search and interactive data exploration.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Kelly, Justin; Jacob, Christian
evoVersion: visualizing evolutionary histories Proceedings Article
In: Evolutionary Computation (CEC), 2016 IEEE Congress on, pp. 814–821, IEEE 2016.
@inproceedings{kelly2016evoversion,
title = {evoVersion: visualizing evolutionary histories},
author = {Justin Kelly and Christian Jacob},
doi = {10.1109/cec.2016.7743875},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
booktitle = {Evolutionary Computation (CEC), 2016 IEEE Congress on},
pages = {814--821},
organization = {IEEE},
abstract = {Evolutionary computation is a field characterized by large data sets produced either through user-driven or automatic evaluation. Because of the sheer magnitude of evolved solutions that need to be reviewed and analyzed, it can be difficult to fully utilize evolutionary data effectively. To address this challenge we present evoVersion, a system capable of applying version control methodologies to assist with both the organization and visualization of evolutionary data for the purpose of building upon previous sessions and assisting with collaborative evolutionary design. Our system is implemented in a 3D game engine, which can make use of immersive and virtual reality visualization of evolutionary design experiments.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
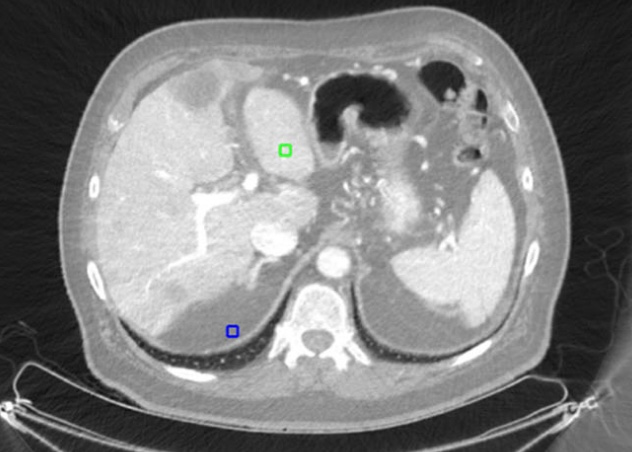
Maia, Rafael Simon; Jacob, Christian; Hara, Amy K; Silva, Alvin C; Pavlicek, William; Mitchell, Ross J
Adaptive noise correction of dual-energy computed tomography images Journal Article
In: International journal of computer assisted radiology and surgery, vol. 11, no. 4, pp. 667–678, 2016.
@article{maia2016adaptive,
title = {Adaptive noise correction of dual-energy computed tomography images},
author = {Rafael Simon Maia and Christian Jacob and Amy K Hara and Alvin C Silva and William Pavlicek and Ross J Mitchell},
doi = {10.1007/s11548-015-1297-8},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
journal = {International journal of computer assisted radiology and surgery},
volume = {11},
number = {4},
pages = {667--678},
publisher = {Springer},
abstract = {Purpose
Noise reduction in material density images is a necessary preprocessing step for the correct interpretation of dual-energy computed tomography (DECT) images. In this paper we describe a new method based on a local adaptive processing to reduce noise in DECT images
Methods
An adaptive neighborhood Wiener (ANW) filter was implemented and customized to use local characteristics of material density images. The ANW filter employs a three-level wavelet approach, combined with the application of an anisotropic diffusion filter. Material density images and virtual monochromatic images are noise corrected with two resulting noise maps.
Results
The algorithm was applied and quantitatively evaluated in a set of 36 images. From that set of images, three are shown here, and nine more are shown in the online supplementary material. Processed images had higher signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) than the raw material density images. The average improvements in SNR and CNR for the material density images were 56.5 and 54.75 %, respectively.
Conclusion
We developed a new DECT noise reduction algorithm. We demonstrate throughout a series of quantitative analyses that the algorithm improves the quality of material density images and virtual monochromatic images.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Noise reduction in material density images is a necessary preprocessing step for the correct interpretation of dual-energy computed tomography (DECT) images. In this paper we describe a new method based on a local adaptive processing to reduce noise in DECT images
Methods
An adaptive neighborhood Wiener (ANW) filter was implemented and customized to use local characteristics of material density images. The ANW filter employs a three-level wavelet approach, combined with the application of an anisotropic diffusion filter. Material density images and virtual monochromatic images are noise corrected with two resulting noise maps.
Results
The algorithm was applied and quantitatively evaluated in a set of 36 images. From that set of images, three are shown here, and nine more are shown in the online supplementary material. Processed images had higher signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) than the raw material density images. The average improvements in SNR and CNR for the material density images were 56.5 and 54.75 %, respectively.
Conclusion
We developed a new DECT noise reduction algorithm. We demonstrate throughout a series of quantitative analyses that the algorithm improves the quality of material density images and virtual monochromatic images.

Yuen, Douglas; Jacob, Christian
Eukaryo: An Agent-based, Interactive Simulation of a Eukaryotic Cell Proceedings Article
In: Proceedings of the Artificial Life Conference 2016, pp. 562–569, MIT Press, Cambridge, MA, 2016.
@inproceedings{Yuen:2016ex,
title = {Eukaryo: An Agent-based, Interactive Simulation of a Eukaryotic Cell},
author = {Douglas Yuen and Christian Jacob},
doi = {10.7551/978-0-262-33936-0-ch090},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
booktitle = {Proceedings of the Artificial Life Conference 2016},
pages = {562--569},
publisher = {MIT Press},
address = {Cambridge, MA},
abstract = {Eukaryo is a 3D, interactive simulation of a eukaryotic cell. In comparison to existing cell simulations, our model illus- trates the structures and processes within a biological cell with increased fidelity and a higher degree of real-time in- teractivity using a virtual reality environment. Implemented in a game engine, Eukaryo is a hybrid model that combines agent-based and mathematical modelling.
Through the use of visual scripting, Eukaryo incorporates both agent-based modelling and mathematical representa- tions to describe gene expression, energy production and waste removal within the cell in a highly visual, interac- tive simulation environment. With the help of virtual real- ity displays, users can be immersed in the crowded spaces of biomolecular worlds and observe metabolic reactions at a high level of detail. Compared to traditional media, such as illustrations and videos, Eukaryo offers superior representa- tions of cellular architecture, its components and dynamics of the machineries of life.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
Through the use of visual scripting, Eukaryo incorporates both agent-based modelling and mathematical representa- tions to describe gene expression, energy production and waste removal within the cell in a highly visual, interac- tive simulation environment. With the help of virtual real- ity displays, users can be immersed in the crowded spaces of biomolecular worlds and observe metabolic reactions at a high level of detail. Compared to traditional media, such as illustrations and videos, Eukaryo offers superior representa- tions of cellular architecture, its components and dynamics of the machineries of life.
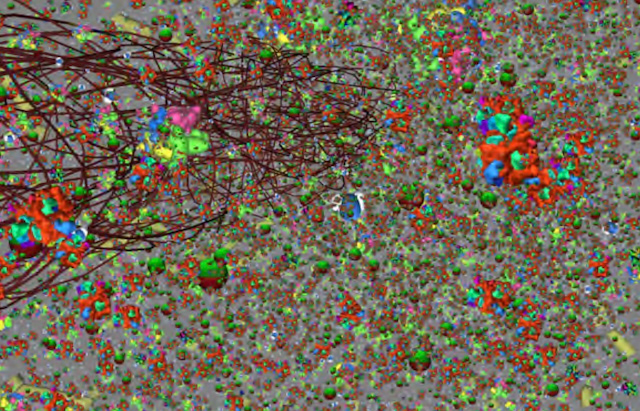
Wu, Andrew; Davison, Timothy; Jacob, Christian
A 3D Multiscale Model of Chemotaxis in Bacteria Proceedings Article
In: Proceedings of the Artificial Life Conference 2016, pp. 546–553, MIT Press, Cambridge, MA, 2016.
@inproceedings{Wu:2016gm,
title = {A 3D Multiscale Model of Chemotaxis in Bacteria},
author = {Andrew Wu and Timothy Davison and Christian Jacob},
doi = {10.7551/978-0-262-33936-0-ch088},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
booktitle = {Proceedings of the Artificial Life Conference 2016},
pages = {546--553},
publisher = {MIT Press},
address = {Cambridge, MA},
abstract = {We present an interactive, agent-based, multi-scale 3D model of a colony of E. coli bacteria. We simulate chemical diffu- sion on an agar plate which is inhabited by a colony of bac- terial cells. The cells interact with a discrete grid that models diffusion of attractants and repellents, to which the cells react. For each bacterium, we simulate its chemotactic behaviour, making a cell either follow a gradient or tumble. Cell propul- sion is determined by the spinning direction of the motors that drive its flagella.
In an agent-based model, we have implemented the molecular elements that comprise the two key chemotactic pathways of excitation and adaptation, which, in turn, regulate the motors and influence a cell’s movement through the agar medium. We show four interconnected model layers that capture the biological processes from the colony layer down to the level of interacting molecules.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
In an agent-based model, we have implemented the molecular elements that comprise the two key chemotactic pathways of excitation and adaptation, which, in turn, regulate the motors and influence a cell’s movement through the agar medium. We show four interconnected model layers that capture the biological processes from the colony layer down to the level of interacting molecules.
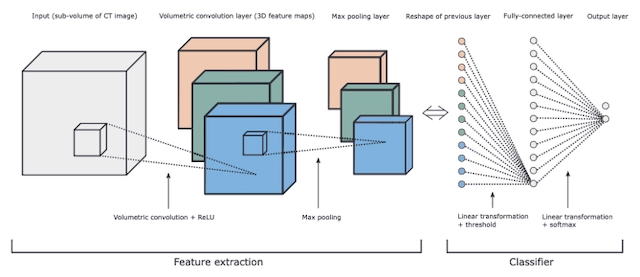
Golan, Rotem; Jacob, Christian; Denzinger, Jörg
Lung nodule detection in CT images using deep convolutional neural networks Proceedings Article
In: Neural Networks (IJCNN), 2016 International Joint Conference on, pp. 243–250, IEEE 2016.
@inproceedings{golan2016lung,
title = {Lung nodule detection in CT images using deep convolutional neural networks},
author = {Rotem Golan and Christian Jacob and Jörg Denzinger},
doi = {10.1109/IJCNN.2016.7727205},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
booktitle = {Neural Networks (IJCNN), 2016 International Joint Conference on},
pages = {243--250},
organization = {IEEE},
abstract = {Early detection of lung nodules in thoracic Computed Tomography (CT) scans is of great importance for the successful diagnosis and treatment of lung cancer. Due to improvements in screening technologies, and an increased demand for their use, radiologists are required to analyze an ever increasing amount of image data, which can affect the quality of their diagnoses. Computer-Aided Detection (CADe) systems are designed to assist radiologists in this endeavor. Here, we present a CADe system for the detection of lung nodules in thoracic CT images. Our system is based on (1) the publicly available Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) database, which contains 1018 thoracic CT scans with nodules of different shape and size, and (2) a deep Convolutional Neural Network (CNN), which is trained, using the back-propagation algorithm, to extract valuable volumetric features from the input data and detect lung nodules in sub-volumes of CT images. Considering only those test nodules that have been annotated by four radiologists, our CADe system achieves a sensitivity (true positive rate) of 78.9% with 20 false positives (FPs) per scan, or a sensitivity of 71.2% with 10 FPs per scan. This is achieved without using any segmentation or additional FP reduction procedures, both of which are commonly used in other CADe systems. Furthermore, our CADe system is validated on a larger number of lung nodules compared to other studies, which increases the variation in their appearance, and therefore, makes their detection by a CADe system more challenging.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Hasan, Mahmudul; Samavati, Faramarz F; Jacob, Christian
Interactive multilevel focus+ context visualization framework Journal Article
In: The Visual Computer, vol. 32, no. 3, pp. 323–334, 2016.
@article{hasan2016interactive,
title = {Interactive multilevel focus+ context visualization framework},
author = {Mahmudul Hasan and Faramarz F Samavati and Christian Jacob},
doi = {10.1007/s00371-015-1180-1},
year = {2016},
date = {2016-01-01},
urldate = {2016-01-01},
journal = {The Visual Computer},
volume = {32},
number = {3},
pages = {323--334},
publisher = {Springer},
abstract = {In this article, we present the construction of an interactive multilevel focus+context visualization framework for the navigation and exploration of large-scale 2D and 3D images. The presented framework utilizes a balanced multiresolution technique supported by a balanced wavelet transform (BWT). It extends the mode of focus+context visualization, where spatially separate magnification of regions of interest (ROIs) is performed, as opposed to in-place magnification. Each resulting visualization scenario resembles a tree structure, where the root constitutes the main context, each non-root internal node plays the dual roles of both focus and context, and each leaf solely represents a focus. Our developed prototype supports interactive manipulation of the visualization hierarchy, such as addition and deletion of ROIs and desired changes in their resolutions at any level of the hierarchy on the fly. We describe the underlying data structure efficiently support such operations. Changes in the spatial locations of query windows defining the ROIs trigger on-demand reconstruction queries. We explain in detail how to efficiently process such reconstruction queries within the hierarchy of details (wavelet coefficients) contained in the BWT in order to ensure real-time feedback. As the BWT need only be constructed once in a preprocessing phase on the server-side and robust on-demand reconstruction queries require minimal data communication overhead, our presented framework is a suitable candidate for efficient web-based visualization of complex large-scale imagery. We also discuss the performance characteristics of our proposed framework from various aspects, such as time and space complexities and achieved frame rates.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2015
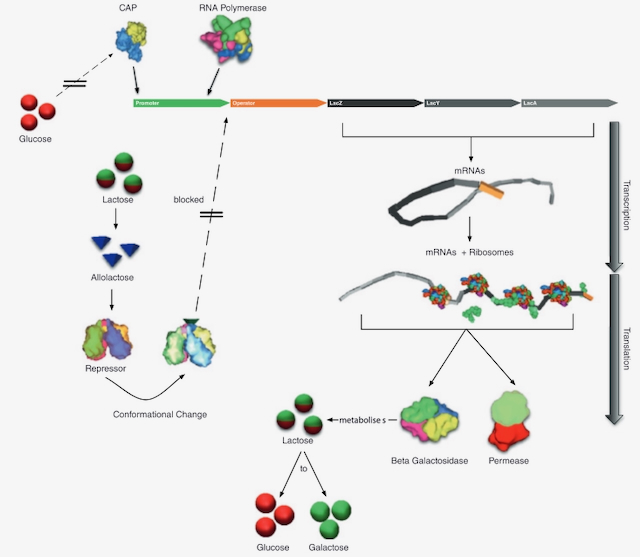
Esmaeili, Afshin; Davison, Timothy; Wu, Andrew; Alcantara, Joenel; Jacob, Christian
PROKARYO: an illustrative and interactive computational model of the lactose operon in the bacterium Escherichia coli Journal Article
In: BMC bioinformatics, vol. 16, no. 1, pp. 311, 2015.
@article{esmaeili2015prokaryo,
title = {PROKARYO: an illustrative and interactive computational model of the lactose operon in the bacterium Escherichia coli},
author = {Afshin Esmaeili and Timothy Davison and Andrew Wu and Joenel Alcantara and Christian Jacob},
doi = {10.1186/s12859-015-0720-z},
year = {2015},
date = {2015-01-01},
urldate = {2015-01-01},
journal = {BMC bioinformatics},
volume = {16},
number = {1},
pages = {311},
publisher = {BioMed Central},
abstract = {Background
We are creating software for agent-based simulation and visualization of bio-molecular processes in bacterial and eukaryotic cells. As a first example, we have built a 3-dimensional, interactive computer model of an Escherichia coli bacterium and its associated biomolecular processes. Our illustrative model focuses on the gene regulatory processes that control the expression of genes involved in the lactose operon. Prokaryo, our agent-based cell simulator, incorporates cellular structures, such as plasma membranes and cytoplasm, as well as elements of the molecular machinery, including RNA polymerase, messenger RNA, lactose permease, and ribosomes.
Results
The dynamics of cellular ’agents’ are defined by their rules of interaction, implemented as finite state machines. The agents are embedded within a 3-dimensional virtual environment with simulated physical and electrochemical properties. The hybrid model is driven by a combination of (1) mathematical equations (DEQs) to capture higher-scale phenomena and (2) agent-based rules to implement localized interactions among a small number of molecular elements. Consequently, our model is able to capture phenomena across multiple spatial scales, from changing concentration gradients to one-on-one molecular interactions.
We use the classic gene regulatory mechanism of the lactose operon to demonstrate our model’s resolution, visual presentation, and real-time interactivity. Our agent-based model expands on a sophisticated mathematical E. coli metabolism model, through which we highlight our model’s scientific validity.
Conclusion
We believe that through illustration and interactive exploratory learning a model system like Prokaryo can enhance the general understanding and perception of biomolecular processes. Our agent-DEQ hybrid modeling approach can also be of value to conceptualize, illustrate, and—eventually—validate cell experiments in the wet lab.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
We are creating software for agent-based simulation and visualization of bio-molecular processes in bacterial and eukaryotic cells. As a first example, we have built a 3-dimensional, interactive computer model of an Escherichia coli bacterium and its associated biomolecular processes. Our illustrative model focuses on the gene regulatory processes that control the expression of genes involved in the lactose operon. Prokaryo, our agent-based cell simulator, incorporates cellular structures, such as plasma membranes and cytoplasm, as well as elements of the molecular machinery, including RNA polymerase, messenger RNA, lactose permease, and ribosomes.
Results
The dynamics of cellular ’agents’ are defined by their rules of interaction, implemented as finite state machines. The agents are embedded within a 3-dimensional virtual environment with simulated physical and electrochemical properties. The hybrid model is driven by a combination of (1) mathematical equations (DEQs) to capture higher-scale phenomena and (2) agent-based rules to implement localized interactions among a small number of molecular elements. Consequently, our model is able to capture phenomena across multiple spatial scales, from changing concentration gradients to one-on-one molecular interactions.
We use the classic gene regulatory mechanism of the lactose operon to demonstrate our model’s resolution, visual presentation, and real-time interactivity. Our agent-based model expands on a sophisticated mathematical E. coli metabolism model, through which we highlight our model’s scientific validity.
Conclusion
We believe that through illustration and interactive exploratory learning a model system like Prokaryo can enhance the general understanding and perception of biomolecular processes. Our agent-DEQ hybrid modeling approach can also be of value to conceptualize, illustrate, and—eventually—validate cell experiments in the wet lab.
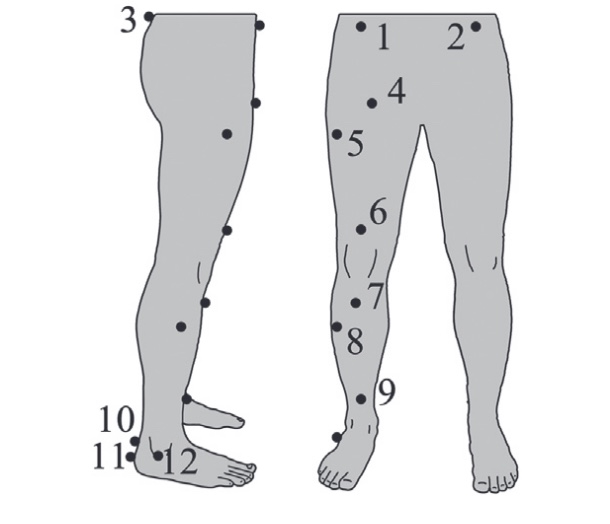
Hoerzer, Stefan; von Tscharner, Vinzenz; Jacob, Christian; Nigg, Benno M
Defining functional groups based on running kinematics using Self-Organizing Maps and Support Vector Machines Journal Article
In: Journal of biomechanics, vol. 48, no. 10, pp. 2072–2079, 2015.
@article{hoerzer2015defining,
title = {Defining functional groups based on running kinematics using Self-Organizing Maps and Support Vector Machines},
author = {Stefan Hoerzer and Vinzenz von Tscharner and Christian Jacob and Benno M Nigg},
doi = {10.1016/j.jbiomech.2015.03.017},
year = {2015},
date = {2015-01-01},
urldate = {2015-01-01},
journal = {Journal of biomechanics},
volume = {48},
number = {10},
pages = {2072--2079},
publisher = {Elsevier},
abstract = {A functional group is a collection of individuals who react in a similar way to a specific intervention/product such as a sport shoe. Matching footwear features to a functional group can possibly enhance footwear-related comfort, improve running performance, and decrease the risk of movement-related injuries. To match footwear features to a functional group, one has to first define the different groups using their distinctive movement patterns. Therefore, the main objective of this study was to propose and apply a methodological approach to define functional groups with different movement patterns using Self-Organizing Maps and Support Vector Machines. Further study objectives were to identify differences in age, gender and footwear-related comfort preferences between the functional groups. Kinematic data and subjective comfort preferences of 88 subjects (16–76 years; 45 m/43 f) were analysed. Eight functional groups with distinctive movement patterns were defined. The findings revealed that most of the groups differed in age or gender. Certain functional groups differed in their comfort preferences and, therefore, had group-specific footwear requirements to enhance footwear-related comfort. Some of the groups, which had group-specific footwear requirements, did not show any differences in age or gender. This is important because when defining functional groups simply using common grouping criteria like age or gender, certain functional groups with group-specific movement patterns and footwear requirements might not be detected. This emphasises the power of the proposed pattern recognition approach to automatically define groups by their distinctive movement patterns in order to be able to address their group-specific product requirements.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
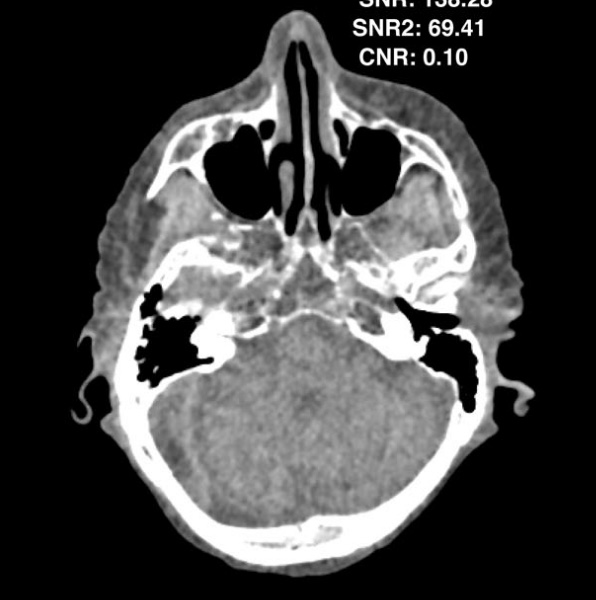
Maia, Rafael Simon; Jacob, Christian; Hara, Amy K; Silva, Alvin C; Pavlicek, William; Ross, Mitchell J
An algorithm for noise correction of dual-energy computed tomography material density images Journal Article
In: International journal of computer assisted radiology and surgery, vol. 10, no. 1, pp. 87–100, 2015.
@article{maia2015algorithm,
title = {An algorithm for noise correction of dual-energy computed tomography material density images},
author = {Rafael Simon Maia and Christian Jacob and Amy K Hara and Alvin C Silva and William Pavlicek and Mitchell J Ross},
doi = {10.1007/s11548-014-1006-z},
year = {2015},
date = {2015-01-01},
urldate = {2015-01-01},
journal = {International journal of computer assisted radiology and surgery},
volume = {10},
number = {1},
pages = {87--100},
publisher = {Springer},
abstract = {Purpose
Dual-energy computed tomography (DECT) images can undergo a two-material decomposition process which results in two images containing material density information. Material density images obtained by that process result in images with increased pixel noise. Noise reduction in those images is desirable in order to improve image quality.
Methods
A noise reduction algorithm for material density images was developed and tested. A three-level wavelet approach combined with the application of an anisotropic diffusion filter was used. During each level, the resulting noise maps are further processed, until the original resolution is reached and the final noise maps obtained. Our method works in image space and, therefore, can be applied to any type of material density images obtained from any DECT vendor. A quantitative evaluation of the noise-reduced images using the signal-to-noise ratio (SNR), contrast-to-noise ratio (CNR) and 2D noise power spectrum was done to quantify the improvements.
Results
The noise reduction algorithm was applied to a set of images resulting in images with higher SNR and CNR than the raw density images obtained by the decomposition process. The average improvement in terms of SNR gain was about 49 % while CNR gain was about 52 %. The difference between the raw and filtered regions of interest mean values was far from reaching statistical significance (minimum p>0.89 , average p>0.97).
Conclusion
We have demonstrated through a series of quantitative analyses that our novel noise reduction algorithm improves the image quality of DECT material density images.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Dual-energy computed tomography (DECT) images can undergo a two-material decomposition process which results in two images containing material density information. Material density images obtained by that process result in images with increased pixel noise. Noise reduction in those images is desirable in order to improve image quality.
Methods
A noise reduction algorithm for material density images was developed and tested. A three-level wavelet approach combined with the application of an anisotropic diffusion filter was used. During each level, the resulting noise maps are further processed, until the original resolution is reached and the final noise maps obtained. Our method works in image space and, therefore, can be applied to any type of material density images obtained from any DECT vendor. A quantitative evaluation of the noise-reduced images using the signal-to-noise ratio (SNR), contrast-to-noise ratio (CNR) and 2D noise power spectrum was done to quantify the improvements.
Results
The noise reduction algorithm was applied to a set of images resulting in images with higher SNR and CNR than the raw density images obtained by the decomposition process. The average improvement in terms of SNR gain was about 49 % while CNR gain was about 52 %. The difference between the raw and filtered regions of interest mean values was far from reaching statistical significance (minimum p>0.89 , average p>0.97).
Conclusion
We have demonstrated through a series of quantitative analyses that our novel noise reduction algorithm improves the image quality of DECT material density images.
2014
Bhalla, Navneet; Bentley, Peter J; Vize, Peter; Jacob, Christian
Staging the self-assembly process: Inspiration from biological development Journal Article
In: Artificial life, vol. 20, no. 1, pp. 29–53, 2014.
@article{bhalla2014staging,
title = {Staging the self-assembly process: Inspiration from biological development},
author = {Navneet Bhalla and Peter J Bentley and Peter Vize and Christian Jacob},
doi = {10.1162/artl_a_00095},
year = {2014},
date = {2014-01-01},
urldate = {2014-01-01},
journal = {Artificial life},
volume = {20},
number = {1},
pages = {29--53},
publisher = {MIT Press},
abstract = {One of the practical challenges facing the creation of self-assembling systems is being able to exploit a limited set of fixed components and their bonding mechanisms. The method of staging divides the self-assembly process into time intervals, during which components can be added to, or removed from, an environment at each interval. Staging addresses the challenge of using components that lack plasticity by encoding the construction of a target structure in the staging algorithm itself and not exclusively in the design of the components. Previous staging strategies do not consider the interplay between component physical features (morphological information). In this work we use morphological information to stage the self-assembly process, during which components can only be added to their environment at each time interval, to demonstrate our concept. Four experiments are presented, which use heterogeneous, passive, mechanical components that are fabricated using 3D printing. Two orbital shaking environments are used to provide energy to the components and to investigate the role of morphological information with component movement in either two or three spatial dimensions. The benefit of our staging strategy is shown by reducing assembly errors and exploiting bonding mechanisms with rotational properties. As well, a doglike target structure is used to demonstrate in theory how component information used at an earlier time interval can be reused at a later time interval, inspired by the use of a body plan in biological development. We propose that a staged body plan is one method toward scaling self-assembling systems with many interacting components. The experiments and body plan example demonstrate, as proof of concept, that staging enables the self-assembly of more complex morphologies not otherwise possible.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Shirazi, Abbas Sarraf; Davison, Timothy; von Mammen, Sebastian; Denzinger, Jörg; Jacob, Christian
Adaptive agent abstractions to speed up spatial agent-based simulations Journal Article
In: Simulation Modelling Practice and Theory, vol. 40, pp. 144–160, 2014.
@article{shirazi2014adaptive,
title = {Adaptive agent abstractions to speed up spatial agent-based simulations},
author = {Abbas Sarraf Shirazi and Timothy Davison and Sebastian von Mammen and Jörg Denzinger and Christian Jacob},
doi = {10.1016/j.simpat.2013.09.001},
year = {2014},
date = {2014-01-01},
urldate = {2014-01-01},
journal = {Simulation Modelling Practice and Theory},
volume = {40},
pages = {144--160},
publisher = {Elsevier},
abstract = {Simulating fine-grained agent-based models requires extensive computational resources. In this article, we present an approach that reduces the number of agents by adaptively abstracting groups of spatial agents into meta-agents that subsume individual behaviours and physical forms. Particularly, groups of agents that have been clustering together for a sufficiently long period of time are detected by observer agents and then abstracted into a single meta-agent. Observers periodically test meta-agents to ensure their validity, as the dynamics of the simulation may change to a point where the individual agents do not form a cluster any more. An invalid meta-agent is removed from the simulation and subsequently, its subsumed individual agents will be put back in the simulation. The same mechanism can be applied on meta-agents thus creating adaptive abstraction hierarchies during the course of a simulation. Experimental results on the simulation of the blood coagulation process show that the proposed abstraction mechanism results in the same system behaviour while speeding up the simulation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
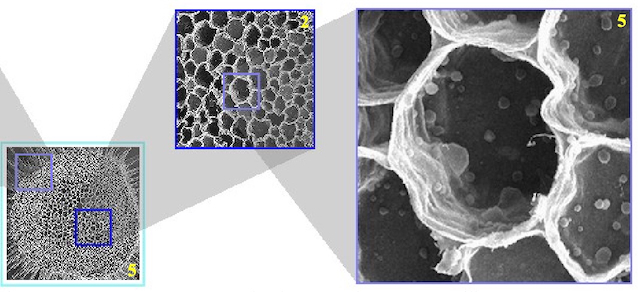
Hasan, Mahmudul; Samavati, Faramarz; Jacob, Christian
Multilevel focus+ context visualization using balanced multiresolution Proceedings Article
In: Cyberworlds (CW), 2014 International Conference on, pp. 145–152, IEEE 2014.
@inproceedings{hasan2014multilevel,
title = {Multilevel focus+ context visualization using balanced multiresolution},
author = {Mahmudul Hasan and Faramarz Samavati and Christian Jacob},
doi = {10.1109/cw.2014.28},
year = {2014},
date = {2014-01-01},
urldate = {2014-01-01},
booktitle = {Cyberworlds (CW), 2014 International Conference on},
pages = {145--152},
organization = {IEEE},
abstract = {In this paper, we present the construction of a multilevel focus context visualization framework for the navigation and exploration of large-scale 2D and 3D images. The presented framework utilizes a balanced multiresolution (BMR) technique supported by a balanced wavelet transform (BWT). This devised framework extends the mode of focus context visualization, where spatially separate magnification of regions of interest (ROIs) is performed, as opposed to in-place magnification. Each resulting visualization scenario resembles a tree structure, where the root constitutes the main context, each non-root internal node plays the dual roles of both focus and context, and each leaf solely represents a focus. We use the local multiresolution filters of quadratic B-spline to construct the BWT. Our developed prototype supports interactive manipulation of the visualization hierarchy, such as addition and deletion of ROIs and desired changes in their resolutions at any level of the hierarchy on the fly. Changes in the spatial locations of query windows that define the ROIs trigger on-demand reconstruction queries. We describe in detail how to efficiently process such reconstruction queries within the hierarchy of details (wavelet coefficients) contained in the BWT in order to ensure real-time feedback. As the BWT need only be constructed once in a preprocessing phase on the server-side and robust on-demand reconstruction queries require minimal data communication overhead, our presented framework is a suitable candidate for efficient web-based visualization and exploration of complex large-scale imagery.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
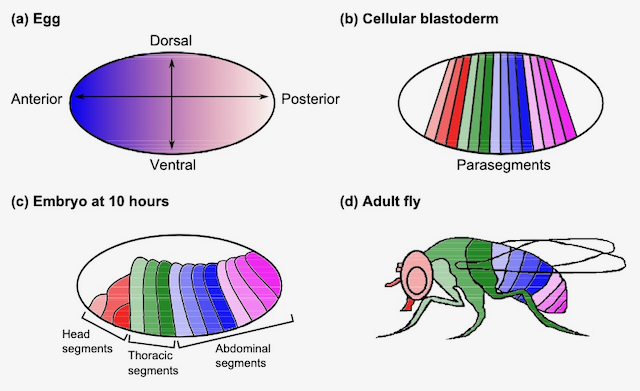
Golan, Rotem; Jacob, Christian; Grewal, Savraj; Denzinger, Jörg
Predicting patterns of gene expression during drosophila embryogenesis Proceedings Article
In: Proceedings of the 2014 Annual Conference on Genetic and Evolutionary Computation, pp. 269–276, ACM 2014.
@inproceedings{golan2014predicting,
title = {Predicting patterns of gene expression during drosophila embryogenesis},
author = {Rotem Golan and Christian Jacob and Savraj Grewal and Jörg Denzinger},
doi = {10.1145/2576768.2598250},
year = {2014},
date = {2014-01-01},
urldate = {2014-01-01},
booktitle = {Proceedings of the 2014 Annual Conference on Genetic and Evolutionary Computation},
pages = {269--276},
organization = {ACM},
abstract = {Understanding how organisms develop from a single cell into a functioning multicellular organism is one of the key questions in developmental biology. Research in this area goes back decades ago, but only recently have improvements in technology allowed biologists to achieve experimental results that are more quantitative and precise. Here, we show how large biological datasets can be used to learn a model for predicting the patterns of gene expression in Drosophila melanogaster (fruit fly) throughout embryogenesis. We also explore the possibility of considering spatial information in order to achieve unique patterns of gene expression in different regions along the anterior-posterior (head-tail) axis of the egg. We then demonstrate how the resulting model can be used to (1) classify these regions into the various segments of the fly, and (2) to conduct a virtual gene knockout experiment. Our learning algorithm is based on a model that has biological meaning, which indicates that its structure and parameters have their correspondence in biology.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
2013
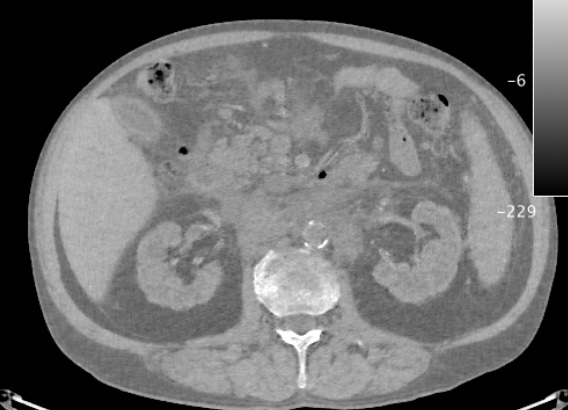
Maia, Rafael Simon; Jacob, Christian; Mitchell, J Ross; Hara, Amy K; Silva, Alvin C; Pavlicek, William
Parallel multi-material decomposition of Dual-Energy CT data Proceedings Article
In: Computer-Based Medical Systems (CBMS), 2013 IEEE 26th International Symposium on, pp. 465–468, IEEE 2013.
@inproceedings{maia2013parallel,
title = {Parallel multi-material decomposition of Dual-Energy CT data},
author = {Rafael Simon Maia and Christian Jacob and J Ross Mitchell and Amy K Hara and Alvin C Silva and William Pavlicek},
doi = {10.1109/cbms.2013.6627842},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
booktitle = {Computer-Based Medical Systems (CBMS), 2013 IEEE 26th International Symposium on},
pages = {465--468},
organization = {IEEE},
abstract = {Dual-Energy Computed Tomography (DECT) is a new modality of CT where two images are acquired simultaneously at two energy levels, and then decomposed into two material density images. It is also possible to further decompose these images into volume fraction images that approximate the percentage of a given material at each pixel. Here, we describe a novel parallel version of the multilateral decomposition algorithm proposed by Mendonça et al., which is used to obtain volume fraction images. Our parallel version accelerates decomposition by 200x. We also discuss some of the algorithm limitations.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
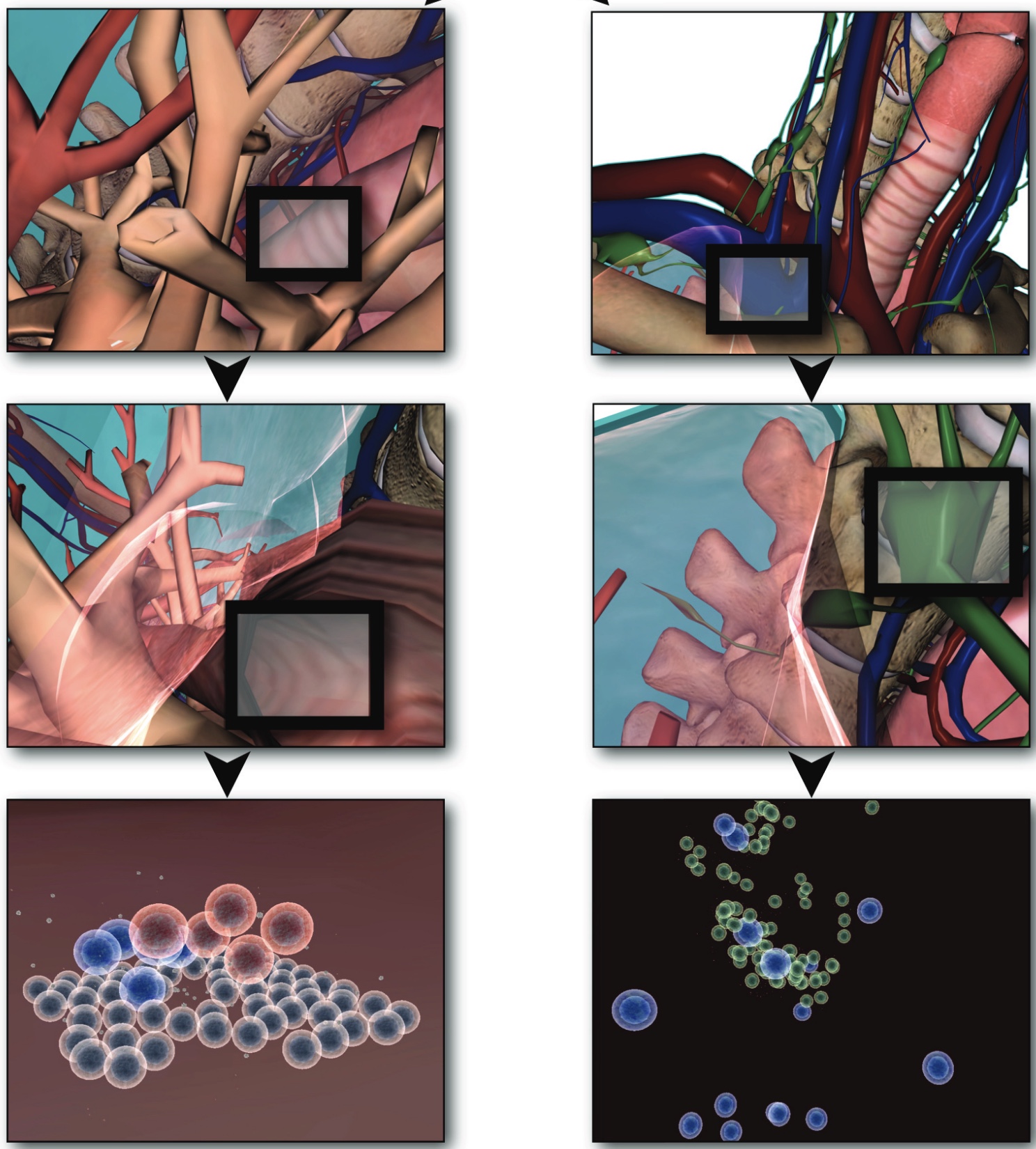
Sarpe, Vladimir; Jacob, Christian
Simulating the decentralized processes of the human immune system in a virtual anatomy model Journal Article
In: BMC bioinformatics, vol. 14, no. 6, pp. S2, 2013.
@article{sarpe2013simulating,
title = {Simulating the decentralized processes of the human immune system in a virtual anatomy model},
author = {Vladimir Sarpe and Christian Jacob},
doi = {10.1186/1471-2105-14-s6-s2},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
journal = {BMC bioinformatics},
volume = {14},
number = {6},
pages = {S2},
publisher = {BioMed Central},
abstract = {Background
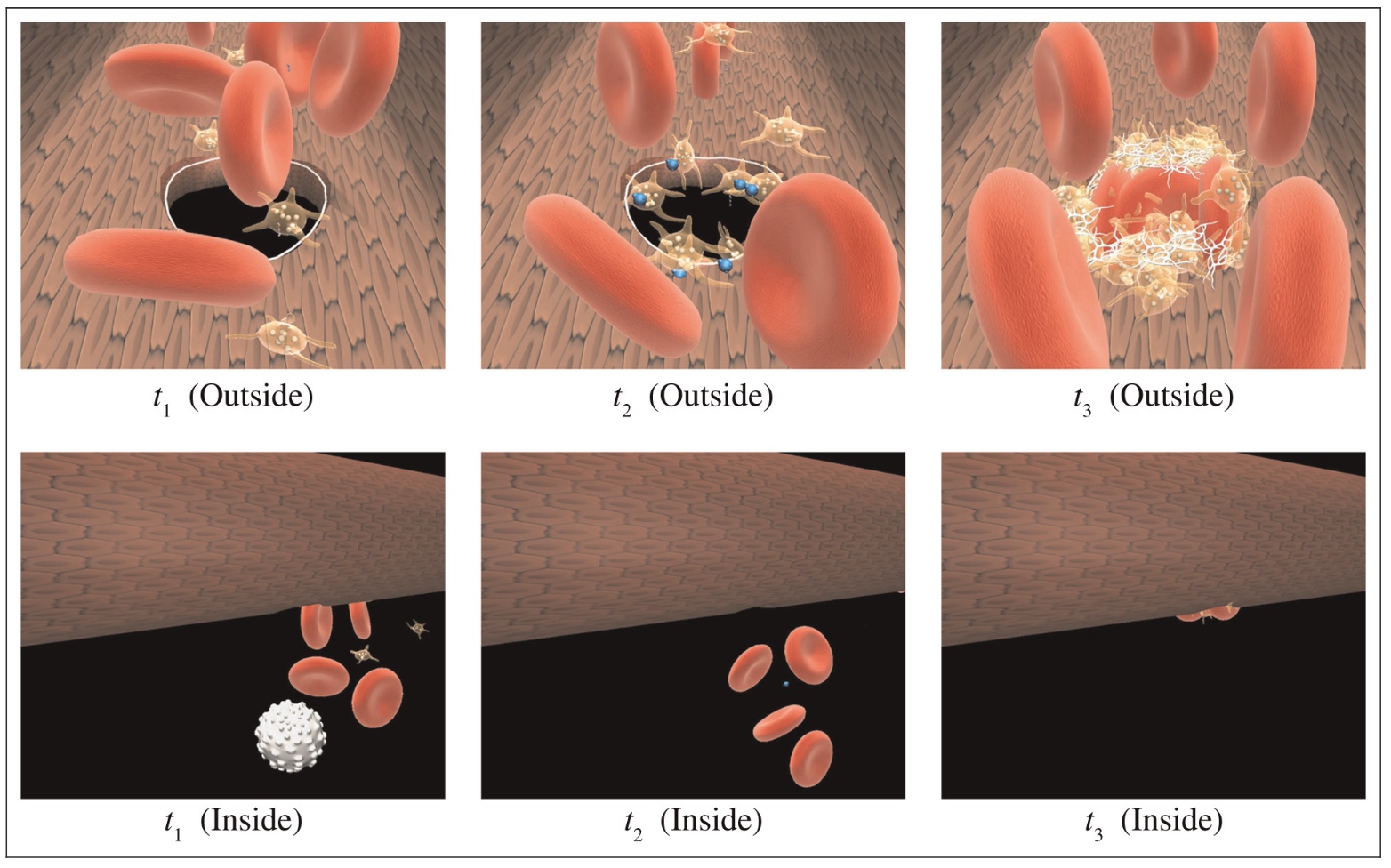
Many physiological processes within the human body can be perceived and modeled as large systems of interacting particles or swarming agents. The complex processes of the human immune system prove to be challenging to capture and illustrate without proper reference to the spacial distribution of immune-related organs and systems. Our work focuses on physical aspects of immune system processes, which we implement through swarms of agents. This is our first prototype for integrating different immune processes into one comprehensive virtual physiology simulation.
Results
Using agent-based methodology and a 3-dimensional modeling and visualization environment (LINDSAY Composer), we present an agent-based simulation of the decentralized processes in the human immune system. The agents in our model - such as immune cells, viruses and cytokines - interact through simulated physics in two different, compartmentalized and decentralized 3-dimensional environments namely, (1) within the tissue and (2) inside a lymph node. While the two environments are separated and perform their computations asynchronously, an abstract form of communication is allowed in order to replicate the exchange, transportation and interaction of immune system agents between these sites. The distribution of simulated processes, that can communicate across multiple, local CPUs or through a network of machines, provides a starting point to build decentralized systems that replicate larger-scale processes within the human body, thus creating integrated simulations with other physiological systems, such as the circulatory, endocrine, or nervous system. Ultimately, this system integration across scales is our goal for the LINDSAY Virtual Human project.
Conclusions
Our current immune system simulations extend our previous work on agent-based simulations by introducing advanced visualizations within the context of a virtual human anatomy model. We also demonstrate how to distribute a collection of connected simulations over a network of computers. As a future endeavour, we plan to use parameter tuning techniques on our model to further enhance its biological credibility. We consider these in silico experiments and their associated modeling and optimization techniques as essential components in further enhancing our capabilities of simulating a whole-body, decentralized immune system, to be used both for medical education and research as well as for virtual studies in immunoinformatics.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Many physiological processes within the human body can be perceived and modeled as large systems of interacting particles or swarming agents. The complex processes of the human immune system prove to be challenging to capture and illustrate without proper reference to the spacial distribution of immune-related organs and systems. Our work focuses on physical aspects of immune system processes, which we implement through swarms of agents. This is our first prototype for integrating different immune processes into one comprehensive virtual physiology simulation.
Results
Using agent-based methodology and a 3-dimensional modeling and visualization environment (LINDSAY Composer), we present an agent-based simulation of the decentralized processes in the human immune system. The agents in our model - such as immune cells, viruses and cytokines - interact through simulated physics in two different, compartmentalized and decentralized 3-dimensional environments namely, (1) within the tissue and (2) inside a lymph node. While the two environments are separated and perform their computations asynchronously, an abstract form of communication is allowed in order to replicate the exchange, transportation and interaction of immune system agents between these sites. The distribution of simulated processes, that can communicate across multiple, local CPUs or through a network of machines, provides a starting point to build decentralized systems that replicate larger-scale processes within the human body, thus creating integrated simulations with other physiological systems, such as the circulatory, endocrine, or nervous system. Ultimately, this system integration across scales is our goal for the LINDSAY Virtual Human project.
Conclusions
Our current immune system simulations extend our previous work on agent-based simulations by introducing advanced visualizations within the context of a virtual human anatomy model. We also demonstrate how to distribute a collection of connected simulations over a network of computers. As a future endeavour, we plan to use parameter tuning techniques on our model to further enhance its biological credibility. We consider these in silico experiments and their associated modeling and optimization techniques as essential components in further enhancing our capabilities of simulating a whole-body, decentralized immune system, to be used both for medical education and research as well as for virtual studies in immunoinformatics.
Tworek, Janet K; Jamniczky, Heather; Jacob, Christian; Hallgrimsson, Benedikt; Wright, Bruce
The LINDSAY Virtual Human Project: An immersive approach to anatomy and physiology Journal Article
In: Anatomical sciences education, vol. 6, no. 1, pp. 19–28, 2013.
@article{tworek2013lindsay,
title = {The LINDSAY Virtual Human Project: An immersive approach to anatomy and physiology},
author = {Janet K Tworek and Heather Jamniczky and Christian Jacob and Benedikt Hallgrimsson and Bruce Wright},
doi = {10.1002/ase.1301},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
journal = {Anatomical sciences education},
volume = {6},
number = {1},
pages = {19--28},
publisher = {Wiley Online Library},
abstract = {The increasing number of digital anatomy teaching software packages challenges anatomy educators on how to best integrate these tools for teaching and learning. Realistically, there exists a complex interplay of design, implementation, politics, and learning needs in the development and integration of software for education, each of which may be further amplified by the somewhat siloed roles of programmers, faculty, and students. LINDSAY Presenter is newly designed software that permits faculty and students to model and manipulate three-dimensional anatomy presentations and images, while including embedded quizzes, links, and text-based content. A validated tool measuring impact across pedagogy, resources, interactivity, freedom, granularity, and factors outside the immediate learning event was used in conjunction with observation, field notes, and focus groups to critically examine the impact of attitudes and perceptions of all stakeholders in the early implementation of LINDSAY Presenter before and after a three-week trial period with the software. Results demonstrate that external, personal media usage, along with students' awareness of the need to apply anatomy to clinical professional situations drove expectations of LINDSAY Presenter. A focus on the software over learning, which can be expected during initial orientation, surprisingly remained after three weeks of use. The time-intensive investment required to create learning content is a detractor from user-generated content and may reflect the consumption nature of other forms of digital learning. Early excitement over new technologies needs to be tempered with clear understanding of what learning is afforded, and how these constructively support future application and integration into professional practice.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Shirazi, Abbas Sarraf; von Mammen, Sebastian; Jacob, Christian
Abstraction of agent interaction processes: Towards large-scale multi-agent models Journal Article
In: Simulation, vol. 89, no. 4, pp. 524–538, 2013.
@article{sarraf2013abstraction,
title = {Abstraction of agent interaction processes: Towards large-scale multi-agent models},
author = {Abbas Sarraf Shirazi and Sebastian von Mammen and Christian Jacob},
doi = {10.1177/0037549712470733},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
journal = {Simulation},
volume = {89},
number = {4},
pages = {524--538},
publisher = {SAGE Publications Sage UK: London, England},
abstract = {The typically large numbers of interactions in agent-based simulations come at considerable computational costs. In this article, we present an approach to reduce the number of interactions based on behavioural patterns that recur during runtime. We employ machine learning techniques to abstract the behaviour of groups of agents to cut down computational complexity while preserving the inherent flexibility of agent-based models. The learned abstractions, which subsume the underlying model agents’ interactions, are constantly tested for their validity: after all, the dynamics of a system may change over time to such an extent that previously learned patterns would not reoccur. An invalid abstraction is, therefore, removed again from the system. The creation and removal of abstractions continues throughout the course of a simulation in order to ensure an adequate adaptation to the system dynamics. Experimental results on biological agent-based simulations show that our proposed approach can successfully reduce the computational complexity during the simulation while maintaining the freedom of arbitrary interactions},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2012

Amorim, Ronan Mendonça; Campos, Ricardo Silva; Lobosco, Marcelo; Jacob, Christian; dos Santos, Rodrigo Weber
An electro-mechanical cardiac simulator based on cellular automata and mass-spring models Book Section
In: Cellular Automata, pp. 434–443, Springer, 2012.
@incollection{amorim2012electro,
title = {An electro-mechanical cardiac simulator based on cellular automata and mass-spring models},
author = {Ronan Mendonça Amorim and Ricardo Silva Campos and Marcelo Lobosco and Christian Jacob and Rodrigo Weber dos Santos},
doi = {10.1007/978-3-642-33350-7_45},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
booktitle = {Cellular Automata},
pages = {434--443},
publisher = {Springer},
abstract = {The mechanical behavior of the heart is guided by the propagation of an electrical wave, called action potential. Many diseases have multiple effects on both electrical and mechanical cardiac physiology. To support a better understanding of the multiscale and multiphysics processes involved in physiological and pathological cardiac conditions, a lot of work has been done in developing computational tools to simulate the electro-mechanical behavior of the heart. In this work, we propose a new user-friendly and efficient tool for the electro-mechanical simulation of the cardiac tissue that is based on cellular automata and mass-spring models. The proposed tool offers a user-friendly interface that allows one to interact with the simulation on-the-fly. In addition, the simulator is parallelized with CUDA and OpenMP to further speedup the execution time of the simulations.},
keywords = {},
pubstate = {published},
tppubtype = {incollection}
}

von Mammen, Sebastian; Phillips, David; Davison, Timothy; Jamniczky, Heather; Hallgrimsson, Benedikt; Jacob, Christian
Swarm-based computational development Book Section
In: Morphogenetic Engineering, pp. 473–499, Springer, 2012.
@incollection{von2012swarm,
title = {Swarm-based computational development},
author = {Sebastian von Mammen and David Phillips and Timothy Davison and Heather Jamniczky and Benedikt Hallgrimsson and Christian Jacob},
doi = {10.1007/978-3-642-33902-8_18},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
booktitle = {Morphogenetic Engineering},
pages = {473--499},
publisher = {Springer},
abstract = {Swarms are a metaphor for complex dynamic systems. In swarms, large numbers of individuals locally interact and form non-linear, dynamic interaction networks. Ants, wasps and termites, for instance, are natural swarms whose individual and group behaviors have been evolving over millions of years. In their intricate nest constructions, the emergent effectiveness of their behaviors becomes apparent. Swarm-based computational simulations capture the corresponding principles of agent-based, decentralized, self-organizing models. In this work, we present ideas around swarm-based developmental systems, in particular swarm grammars, a swarm-based generative representation, and our efforts towards the unification of this methodology and the improvement of its accessibility.},
keywords = {},
pubstate = {published},
tppubtype = {incollection}
}

Jacob, Christian; von Mammen, Sebastian; Davison, Timothy; Sarraf-Shirazi, Abbas; Sarpe, Vladimir; Esmaeili, Afshin; Phillips, David; Yazdanbod, Iman; Novakowski, Scott; Steil, Scott; Gingras, Carey; Jamniczky, Heather; Hallgrimsson, Benedikt; Wright, Bruce
Lindsay virtual human: Multi-scale, agent-based, and interactive Book Section
In: Advances in Intelligent Modelling and Simulation, pp. 327–349, Springer, 2012.
@incollection{jacob2012lindsay,
title = {Lindsay virtual human: Multi-scale, agent-based, and interactive},
author = {Christian Jacob and Sebastian von Mammen and Timothy Davison and Abbas Sarraf-Shirazi and Vladimir Sarpe and Afshin Esmaeili and David Phillips and Iman Yazdanbod and Scott Novakowski and Scott Steil and Carey Gingras and Heather Jamniczky and Benedikt Hallgrimsson and Bruce Wright},
doi = {10.1007/978-3-642-30154-4_14},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
booktitle = {Advances in Intelligent Modelling and Simulation},
pages = {327--349},
publisher = {Springer},
abstract = {We are developing LINDSAY Virtual Human, a 3-dimensional, interactive computer model of male and female anatomy and physiology. LINDSAY is designed to be used for medical education. One key characteristic of LINDSAY is the integration of computational models across a range of spatial and temporal scales. We simulate physiological processes in an integrative fashion: from the body level to the level of organs, tissues, cells, and sub-cellular structures. For use in the classroom, we have built LINDSAY Presenter, a 3D slide-based visualization and exploration environment that presents different scenarios within the simulated human body. We are developing LINDSAY Composer to create complex scenes for demonstration, exploration and investigation of physiological scenarios. At LINDSAY Composer′s core is a graphical programming environment, which facilitates the composition of complex, interactive educational modules around the human body.},
keywords = {},
pubstate = {published},
tppubtype = {incollection}
}

von Mammen, Sebastian; Shirazi, Abbas Sarraf; Sarpe, Vladimir; Jacob, Christian
Optimization of swarm-based simulations Journal Article
In: ISRN Artificial Intelligence, vol. 2012, 2012.
@article{von2012optimization,
title = {Optimization of swarm-based simulations},
author = {Sebastian von Mammen and Abbas Sarraf Shirazi and Vladimir Sarpe and Christian Jacob},
doi = {10.5402/2012/365791},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
journal = {ISRN Artificial Intelligence},
volume = {2012},
publisher = {Hindawi Publishing Corporation},
abstract = {In computational swarms, large numbers of reactive agents are simulated. The swarm individuals may coordinate their movements in a “search space” to create efficient routes, to occupy niches, or to find the highest peaks. From a more general perspective though, swarms are a means of representation and computation to bridge the gap between local, individual interactions, and global, emergent phenomena. Computational swarms bear great advantages over other numeric methods, for instance, regarding their extensibility, potential for real-time interaction, dynamic interaction topologies, close translation between natural science theory and the computational model, and the integration of multiscale and multiphysics aspects. However, the more comprehensive a swarm-based model becomes, the more demanding its configuration and the more costly its computation become. In this paper, we present an approach to effectively configure and efficiently compute swarm-based simulations by means of heuristic, population-based optimization techniques. We emphasize the commonalities of several of our recent studies that shed light on top-down model optimization and bottom-up abstraction techniques, culminating in a postulation of a general concept of self-organized optimization in swarm-based simulations.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Bhalla, Navneet; Bentley, Peter J; Vize, Peter D; Jacob, Christian
Programming and evolving physical self-assembling systems in three dimensions Journal Article
In: Natural Computing, vol. 11, no. 3, pp. 475–498, 2012.
@article{bhalla2012programming,
title = {Programming and evolving physical self-assembling systems in three dimensions},
author = {Navneet Bhalla and Peter J Bentley and Peter D Vize and Christian Jacob},
doi = {10.1007/s11047-011-9293-6},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
journal = {Natural Computing},
volume = {11},
number = {3},
pages = {475--498},
publisher = {Springer},
abstract = {Being able to engineer a set of components and their corresponding environmental conditions such that target entities emerge as the result of self-assembly remains an elusive goal. In particular, understanding how to exploit physical properties to create self-assembling systems in three dimensions (in terms of component movement) with symmetric and asymmetric features is extremely challenging. Furthermore, primarily top-down design methodologies have been used to create physical self-assembling systems. As the sophistication of these systems increases, it will be more challenging to use top-down design due to self-assembly being an algorithmically NP-complete problem. In this work, we first present a nature-inspired approach to demonstrate how physically encoded information can be used to program and direct the self-assembly process in three dimensions. Second, we extend our nature-inspired approach by incorporating evolutionary computing, to couple bottom-up construction (self-assembly) with bottom-up design (evolution). To demonstrate our design approach, we present eight proof-of-concept experiments where virtual component sets either defined (programmed) or generated (evolved) during the design process have their specifications translated and fabricated using rapid prototyping. The resulting mechanical components are placed in a jar of fluid on an orbital shaker, their environment. The energy and physical properties of the environment, along with the physical properties of the components (including complementary shapes and magnetic-bit patterns, created using permanent magnets to attract and repel components) are used to engineer the self-assembly process to create emergent target structures with three-dimensional symmetric and asymmetric features. The successful results demonstrate how physically encoded information can be used with programming and evolving physical self-assembling systems in three dimensions.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2011

Bhalla, Navneet; Bentley, Peter J; Vice, Peter; Jacob, Christian
Staging the Self-Assembly Process Using Morphological Information Conference
ECAL 2011 - 20th European Conference on Artificial Life, Paris, France, 2011.
@conference{280,
title = {Staging the Self-Assembly Process Using Morphological Information},
author = {Navneet Bhalla and Peter J Bentley and Peter Vice and Christian Jacob},
url = {https://mitpress.mit.edu/sites/default/files/titles/alife/0262297140chap18.pdf},
year = {2011},
date = {2011-08-01},
urldate = {2011-08-01},
booktitle = {ECAL 2011 - 20th European Conference on Artificial Life},
address = {Paris, France},
abstract = {One of the practical challenges facing the creation of self- assembling systems is being able to exploit a limited set of fixed components and their bonding mechanisms. Staging ad- dresses this challenge by dividing the self-assembly process into time intervals, and encodes the construction of a target structure in the staging algorithm itself and not exclusively into the design of components. Previous staging strategies do not consider the interplay between component physical fea- tures (morphological information). In this work we use mor- phological information to stage the self-assembly process, with the benefit of reducing assembly errors and leveraging bonding mechanism with rotational properties. Four experi- ments are presented, which use heterogeneous, passive, me- chanical components that are fabricated using rapid prototyp- ing. Two orbital shaking environments are used to provide en- ergy to the components, and to investigate the role of morpho- logical information with component movement in either two or three spatial dimensions. The experiments demonstrate, as proof-of-concept, that staging enables the self-assembly of more complex morphologies not otherwise possible.},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}

Shirazi, Abbas Sarraf; von Mammen, Sebastian; Yazdanbod, Iman; Jacob, Christian
Self-Organized Learning of Collective Behaviours in Agent-Based Simulations Conference
ICCS 2011 - 8th International Conference on Complex Systems, NECSI Knowledge Press NECSI Knowledge Press, Boston, MA, USA, 2011, ISBN: 978-0-9656328-4-3.
@conference{282,
title = {Self-Organized Learning of Collective Behaviours in Agent-Based Simulations},
author = {Abbas Sarraf Shirazi and Sebastian von Mammen and Iman Yazdanbod and Christian Jacob},
url = {http://s3.amazonaws.com/academia.edu.documents/5575416/284.pdf?AWSAccessKeyId=AKIAIWOWYYGZ2Y53UL3A&Expires=1493413302&Signature=aRsF8s9E3774YmTdjMOYnAEHdn4%3D&response-content-disposition=inline%3B%20filename%3DSelf-Organized_Learning_of_Collective_Be.pdf},
isbn = {978-0-9656328-4-3},
year = {2011},
date = {2011-06-01},
urldate = {2011-06-01},
booktitle = {ICCS 2011 - 8th International Conference on Complex Systems},
publisher = {NECSI Knowledge Press},
address = {Boston, MA, USA},
organization = {NECSI Knowledge Press},
abstract = {<p>In this paper, we propose an approach to reduce the number of interactions among agents in a multi-agent simulation. Modeling agent interactions turns out to be com- putationally expensive, especially when arbitrary interactions are allowed. In order to diminish computational costs of simulating agent interactions, we propose a self- organized approach to abstracting recurrent interaction patterns among groups of agents. Predictably interacting groups of agents are subsumed by higher-order agents that reproduce similar behaviours but at reduced computational costs. To this end, ob- server agents are immersed into the simulation space in order to monitor groups of agents and learn interaction patterns. Since the dynamics of the system changes over time, an abstraction might loose its validity and must therefore be removed again. This process is regulated by confidence values that are calculated and associated with individual abstractions. If a pattern exists for longer than anticipated, its confidence value is increased. The process of creating and removing abstractions is repeated dur- ing the course of a simulation in order to ensure an adequate adaptation to the system dynamics. Experimental results on a biological agent-based simulation show that our proposed abstraction method can successfully reduce the computational complexity during the simulation while maintaining the possibility of arbitrary interactions.</p>
<p>~</p> <p><img src="webkit-fake-url://414BFFCE-977B-4593-895A-21E7D4530D66/image.tiff" /></p>},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}
<p>~</p> <p><img src="webkit-fake-url://414BFFCE-977B-4593-895A-21E7D4530D66/image.tiff" /></p>

Tsang, Herbert H; Jacob, Christian
RNADPCompare: an algorithm for comparing RNA secondary structures based on image processing techniques Proceedings Article
In: Evolutionary Computation (CEC), 2011 IEEE Congress on, pp. 1288–1295, IEEE 2011.
@inproceedings{tsang2011rnadpcompare,
title = {RNADPCompare: an algorithm for comparing RNA secondary structures based on image processing techniques},
author = {Herbert H Tsang and Christian Jacob},
doi = {10.1109/cec.2011.5949764},
year = {2011},
date = {2011-01-01},
urldate = {2011-01-01},
booktitle = {Evolutionary Computation (CEC), 2011 IEEE Congress on},
pages = {1288--1295},
organization = {IEEE},
abstract = {In structural biology, structural chemistry, and bioinformatics, Ribonucleic Acid (RNA) structure comparison is a fundamental problem. It is because structural comparison can facilitate RNA structure prediction and studies in RNA energy landscapes and conformational switches as well. There are many different tools have been proposed for RNA secondary structure comparison. This paper describes and presents a novel algorithm, RNADPCompare, for computing similarity measure of RNA secondary structures. The main idea for this algorithm is to represent the RNA secondary structure as a dot plot, and then process the dot plot as an image. The algorithm will utilize image processing techniques and heuristic understanding of the image properties to compute similarity measure of RNA secondary structures. Since many evolutionary and machine learning algorithms for RNA secondary structure design and prediction rely on good metric for examining structural similarities, therefore this novel metric will make significant contribution to the advances to these algorithms. An evaluation of the algorithm in terms of correlation to the native structure is made. The results from the six sequences of RNA from a variety of sequence lengths and organisms were tested. When comparing with Sfold, the prediction accuracy of using RNADPCompare to compute the difference matrix seems to be very promising. These results demonstrated that RNADPCompare is highly competitive in terms of the processing speed and accuracy when compare to other methods. This supports the use of this algorithm on other research in RNA secondary structure design and prediction.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Shirazi, Abbas Sarraf; von Mammen, Sebastian; Jacob, Christian
Hierarchical self-organized learning in agent-based modeling of the MAPK signaling pathway Proceedings Article
In: Evolutionary Computation (CEC), 2011 IEEE Congress on, pp. 2245–2251, IEEE 2011.
@inproceedings{shirazi2011hierarchical,
title = {Hierarchical self-organized learning in agent-based modeling of the MAPK signaling pathway},
author = {Abbas Sarraf Shirazi and Sebastian von Mammen and Christian Jacob},
doi = {10.1109/cec.2011.5949893},
year = {2011},
date = {2011-01-01},
urldate = {2011-01-01},
booktitle = {Evolutionary Computation (CEC), 2011 IEEE Congress on},
pages = {2245--2251},
organization = {IEEE},
abstract = {In this paper, we present a self-organized approach to automatically identify and create hierarchies of cooperative agents. Once a group of cooperative agents is found, a higher order agent is created which in turn learns the group behaviour. This way, the number of agents and thus the complexity of the multiagent system will be reduced, as one agent emulates the behaviour of several agents. Our proposed method of creating hierarchies captures the dynamics of a multiagent system by adaptively creating and breaking down hierarchies of agents as the simulation proceeds. Experimental results on two MAPK signaling pathways suggest that the proposed approach is suitable in stable systems while periodic systems still need further investigations.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

von Mammen, Sebastian; Steghöfer, Jan-Philipp; Denzinger, Jörg; Jacob, Christian
Self-organized middle-out abstraction Proceedings Article
In: International Workshop on Self-Organizing Systems, pp. 26–31, Springer 2011.
@inproceedings{von2011self,
title = {Self-organized middle-out abstraction},
author = {Sebastian von Mammen and Jan-Philipp Steghöfer and Jörg Denzinger and Christian Jacob},
doi = {10.1007/978-3-642-19167-1_3},
year = {2011},
date = {2011-01-01},
urldate = {2011-01-01},
booktitle = {International Workshop on Self-Organizing Systems},
pages = {26--31},
organization = {Springer},
abstract = {In this position paper we present a concept to automatically simplify computational processes in large-scale self-organizing multi-agent simulations. The fundamental idea is that groups of agents that exhibit predictable interaction patterns are temporarily subsumed by higher order agents with behaviours of lower computational costs. In this manner, hierarchies of meta-agents automatically abstract large-scale systems involving agents with in-depth behavioural descriptions, rendering the process of upfront simplification obsolete that is usually necessary in numerical approaches. Abstraction hierarchies are broken down again as soon as they become invalid, so that the loss of valuable process information due to simplification is minimized. We describe the algorithm and the representation, we argue for its general applicability and potential power and we underline the challenges that will need to be overcome.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Jacob, Christian; Sarpe, Vladimir; Gingras, Carey; Feyt, Rolando Pajon
Swarm-based simulations for immunobiology Book Section
In: Information Processing and Biological Systems, pp. 29–64, Springer, 2011.
@incollection{jacob2011swarm,
title = {Swarm-based simulations for immunobiology},
author = {Christian Jacob and Vladimir Sarpe and Carey Gingras and Rolando Pajon Feyt},
doi = {10.1007/978-3-642-19621-8_3},
year = {2011},
date = {2011-01-01},
urldate = {2011-01-01},
booktitle = {Information Processing and Biological Systems},
pages = {29--64},
publisher = {Springer},
abstract = {In this contribution, we present a computer model of information processing within a highly distributed biological system of the human body, which is orchestrated over multiple scales of time and space: the immune system. We consider the human body and its environment as a well-orchestrated system of interacting swarms: swarms of cells, swarms of messenger molecules, swarms of bacteria, and swarms of viruses. Utilizing swarm intelligence techniques, we present three virtual simulations and experiments to explore key aspects of the human immune system. Immune system cells and related entities (viruses, bacteria, cytokines) are represented as virtual agents inside 3-dimensional, decentralized compartments that represent primary and secondary lymphoid organs as well as vascular and lymphatic vessels. Specific immune system responses emerge as by-products from collective interactions among the involved simulated ‘agents’ and their environment. We demonstrate simulation results for clonal selection in combination with primary and secondary collective responses after viral infection. We also model, simulate, and visualize key response patterns encountered during bacterial infection. As a third model we consider the complement system, for which we present initial simulation results. We consider these in-silico experiments and their associated modeling environments as an essential step towards hierarchical whole-body simulations of the immune system, both for educational and research purposes.},
keywords = {},
pubstate = {published},
tppubtype = {incollection}
}

von Mammen, Sebastian; Wißmeier, Thomas; Wong, Joyce; Jacob, Christian
Artistic exploration of the worlds of digital developmental swarms Journal Article
In: Leonardo, vol. 44, no. 1, pp. 5–13, 2011.
@article{von2011artistic,
title = {Artistic exploration of the worlds of digital developmental swarms},
author = {Sebastian von Mammen and Thomas Wißmeier and Joyce Wong and Christian Jacob},
doi = {10.1162/leon_a_00087},
year = {2011},
date = {2011-01-01},
urldate = {2011-01-01},
journal = {Leonardo},
volume = {44},
number = {1},
pages = {5--13},
publisher = {MIT Press},
abstract = {This paper presents artwork that was inspired by a computational model called Swarm Grammars. In this work, the “liveliness” of swarms is combined with the generative capabilities of more established developmental representations. Three of the authors followed their individual artistic approaches to explore the creativity and dynamics of Swarm Grammar structures. One chose to breed structures interactively to compose virtual spaces. The second explores the movement and construction dynamics of interactive swarms. The third artist translated developmental processes of Swarm Grammars into interactions of paint particles driven by friction and gravity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2010

Pilat, Marcin; Jacob, Christian
Evolution of Vision Capabilities in Embodied Virtual Creatures Conference
GECCO 2010: Genetic and Evolutionary Computation Conference, Portland, Oregon, USA, 2010.
@conference{250,
title = {Evolution of Vision Capabilities in Embodied Virtual Creatures},
author = {Marcin Pilat and Christian Jacob},
doi = {10.1145/1830483.1830502},
year = {2010},
date = {2010-07-01},
urldate = {2010-07-01},
booktitle = {GECCO 2010: Genetic and Evolutionary Computation Conference},
address = {Portland, Oregon, USA},
abstract = {We evolve light following behaviours in virtual creatures through neural network training using an incremental evolution approach. The neural controllers of creatures evolved for movement are augmented with simple visual neurons and neural connections. Using an evolutionary algorithm, the resulting creatures are trained to identify and follow a light source. Through this process, we are able to train the neural controllers to create various light following behaviours. Many of the evolved behaviours show stability and adaptiveness to environmental perturbations of body orientation.},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}

von Mammen, Sebastian; Davison, Timothy; Baghi, Hamid; Jacob, Christian
Component-Based Networking for Simulations in Medical Education Conference
MediaWiN 2010: 5th IEEE Workshop on multiMedia Applications over Wireless Networks, Ricione, Italy, 2010.
@conference{247,
title = {Component-Based Networking for Simulations in Medical Education},
author = {Sebastian von Mammen and Timothy Davison and Hamid Baghi and Christian Jacob},
doi = {10.1109/iscc.2010.5546594},
year = {2010},
date = {2010-06-01},
urldate = {2010-06-01},
booktitle = {MediaWiN 2010: 5th IEEE Workshop on multiMedia Applications over Wireless Networks},
address = {Ricione, Italy},
abstract = {For the purpose of medical education, our research team is creating LINDSAY, a 3-dimensional, interactive computer model of male and female anatomy and physiology. As part of the LINDSAY—Virtual Human project, we have developed a component-based computational framework that allows the utilization of various formal representations, computation engines and visualization technologies within a single simulation context. For our agent-based simulations, the graphics, physics and behaviours of our interacting entities are implemented through a set of component engines. We have developed a light-weight client/server component, which spreads its siblings in the system's component hierarchy over a wireless or wired network infrastructure. In this paper we demonstrate how our client/server component paves new ways for organizing, generating, computing and presenting educational contents.},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}

Sarpe, Vladimir; Esmaeili, Afshin; Yazdanbod, Iman; Kubik, Thane; Richter, Michael; Jacob, Christian
Parametric evolution of a bacterial signalling system formalized by membrane computing Proceedings Article
In: Evolutionary Computation (CEC), 2010 IEEE Congress on, pp. 1–8, IEEE 2010.
@inproceedings{sarpe2010parametric,
title = {Parametric evolution of a bacterial signalling system formalized by membrane computing},
author = {Vladimir Sarpe and Afshin Esmaeili and Iman Yazdanbod and Thane Kubik and Michael Richter and Christian Jacob},
doi = {10.1109/cec.2010.5585992},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {Evolutionary Computation (CEC), 2010 IEEE Congress on},
pages = {1--8},
organization = {IEEE},
abstract = {Abstract:
In this work, we use particle swarm optimization (PSO) to adjust the parameters of a membrane computing (MC) model of a synthetic autoinducer-2 (AI-2) signalling system in genetically engineered Escherichia coli bacteria. Bacteria release, receive and recognize signalling molecules in order to exchange information. These signalling molecules are responsible for coordinating gene expression at the population level in response to various stimuli such as size of the population, nutrient availability and other biochemical signals. This bacterial cell-to-cell communication is known as Quorum Sensing (QS). AI-2, from Vibrio harveyi, is the signaling molecule of interest in this study. We present a non-deterministic in silico model of Autoinducer-2 Quorum Sensing that is formalized by membrane computing (MC). The model is driven by 23 interaction rules that define biochemical interactions between independent compartments known as membranes. Due to the high dimensionality of this problem as well as lack of data relating to the biochemical parameters of this signalling system, we used a generic particle swarm optimization (PSO) algorithm to discover optimal solutions for the rule stochasticity constants. Our results were compared to the expected trends in quorum sensing behaviour. Ultimately, the results obtained from the PSO are thought to be in accordance with the predicted behaviour of the synthetic AI-2 signalling system.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
In this work, we use particle swarm optimization (PSO) to adjust the parameters of a membrane computing (MC) model of a synthetic autoinducer-2 (AI-2) signalling system in genetically engineered Escherichia coli bacteria. Bacteria release, receive and recognize signalling molecules in order to exchange information. These signalling molecules are responsible for coordinating gene expression at the population level in response to various stimuli such as size of the population, nutrient availability and other biochemical signals. This bacterial cell-to-cell communication is known as Quorum Sensing (QS). AI-2, from Vibrio harveyi, is the signaling molecule of interest in this study. We present a non-deterministic in silico model of Autoinducer-2 Quorum Sensing that is formalized by membrane computing (MC). The model is driven by 23 interaction rules that define biochemical interactions between independent compartments known as membranes. Due to the high dimensionality of this problem as well as lack of data relating to the biochemical parameters of this signalling system, we used a generic particle swarm optimization (PSO) algorithm to discover optimal solutions for the rule stochasticity constants. Our results were compared to the expected trends in quorum sensing behaviour. Ultimately, the results obtained from the PSO are thought to be in accordance with the predicted behaviour of the synthetic AI-2 signalling system.

Tsang, Herbert H; Jiang, Tiancheng; Wiese, Kay C; Jacob, Christian
A study of rna secondary structure prediction using different mutation operators Proceedings Article
In: Evolutionary Computation (CEC), 2010 IEEE Congress on, pp. 1–7, IEEE 2010.
@inproceedings{tsang2010study,
title = {A study of rna secondary structure prediction using different mutation operators},
author = {Herbert H Tsang and Tiancheng Jiang and Kay C Wiese and Christian Jacob},
doi = {10.1109/cec.2010.5585923},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {Evolutionary Computation (CEC), 2010 IEEE Congress on},
pages = {1--7},
organization = {IEEE},
abstract = {Ribonucleic Acid (RNA) has important structural and functional roles in the cell and plays roles in many stages of protein synthesis as well. The functions of RNA molecules are determined largely by their three-dimensional structure. SARNA-Predict has shown excellent results in predicting RNA secondary structure based on Simulated Annealing (SA). SARNA-Predict uses a permutation-based representation to the RNA secondary structure and this paper investigates the impact of the mutation operators in this algorithm. Experiments were performed using a sample of eleven sequences from four RNA classes. The results presented in this paper demonstrate that SARNA-Predict using the percentage swap translocating mutation operator can produce similar results when compared with previous research. Furthermore, the new operator has the potential of reaching a solution with a lower free energy. This supports the use of the proposed operator on RNA secondary structure prediction of other known structures.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}
Bhalla, Navneet; Bentley, Peter J; Jacob, Christian
Evolving physical self-assembling systems in two-dimensions Proceedings Article
In: International Conference on Evolvable Systems, pp. 381–392, Springer 2010.
@inproceedings{bhalla2010evolving,
title = {Evolving physical self-assembling systems in two-dimensions},
author = {Navneet Bhalla and Peter J Bentley and Christian Jacob},
doi = {10.1007/978-3-642-15323-5_33},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {International Conference on Evolvable Systems},
pages = {381--392},
organization = {Springer},
abstract = {Primarily top-down design methodologies have been used to create physical self-assembling systems. As the sophistication of these systems increases, it will be more challenging to deploy top-down design, due to self-assembly being an algorithmically NP-complete problem. Alternatively, we present a nature-inspired approach incorporating evolutionary computing, to couple bottom-up construction (self-assembly) with bottom-up design (evolution). We also present two experiments where evolved virtual component sets are fabricated using rapid prototyping and placed on the surface of an orbital shaking tray, their environment. The successful results demonstrate how this approach can be used for evolving physical self-assembling systems in two-dimensions.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Davison, Timothy; von Mammen, Sebastian; Jacob, Christian
Evoshelf: a system for managing and exploring evolutionary data Proceedings Article
In: International Conference on Parallel Problem Solving from Nature, pp. 310–319, Springer 2010.
@inproceedings{davison2010evoshelf,
title = {Evoshelf: a system for managing and exploring evolutionary data},
author = {Timothy Davison and Sebastian von Mammen and Christian Jacob},
doi = {10.1007/978-3-642-15871-1_32},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {International Conference on Parallel Problem Solving from Nature},
pages = {310--319},
organization = {Springer},
abstract = {Systems that utilize evolutionary computation produce large amounts of data. Quite often, this data has a convenient visual representation. However, managing and visualizing evolutionary data can be a difficult and onerous task. By employing techniques used in photo management software, we have produced a system that helps to organize and visualize evolutionary data while retaining a complete record of a simulation. By means of a simple plugin architecture this system can be extended to import data produced by arbitrary evolutionary systems. We present the system’s architecture, its features, and we provide a comprehensive example, highlighting its advantages in applied research.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Shirazi, Abbas Sarraf; von Mammen, Sebastian; Jacob, Christian
Adaptive modularization of the MAPK signaling pathway using the multiagent paradigm Proceedings Article
In: International Conference on Parallel Problem Solving from Nature, pp. 401–410, Springer 2010.
@inproceedings{shirazi2010adaptive,
title = {Adaptive modularization of the MAPK signaling pathway using the multiagent paradigm},
author = {Abbas Sarraf Shirazi and Sebastian von Mammen and Christian Jacob},
doi = {10.1007/978-3-642-15871-1_41},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {International Conference on Parallel Problem Solving from Nature},
pages = {401--410},
organization = {Springer},
abstract = {We utilize an agent-based approach to model the MAPK signaling pathway, in which we capture both individual and group behaviour of the biological entities inside the system. In an effort to adaptively reduce complexity of interactions among the simulated agents, we propose a bottom-up approach to find and group similar agents into a single module which will result in a reduction in the complexity of the system. Our proposed adaptive method of grouping and ungrouping captures the dynamics of the system by identifying and breaking modules adaptively as the simulation proceeds. Experimental results on our simulated MAPK signaling pathway show that our proposed method can be used to identify modules in both stable and periodic systems.},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}

Khemka, Namrata; Jacob, Christian
VISPLORE: Exploring Particle Swarms by Visual Inspection Book Section
In: Agent-Based Evolutionary Search, pp. 255–284, Springer, 2010.
@incollection{khemka2010visplore,
title = {VISPLORE: Exploring Particle Swarms by Visual Inspection},
author = {Namrata Khemka and Christian Jacob},
doi = {10.1007/978-3-642-13425-8_12},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {Agent-Based Evolutionary Search},
pages = {255--284},
publisher = {Springer},
abstract = {We describe VISPLORE, a visualization and experimentation environment for population-based search algorithms. Using particle swarm optimization (PSO) as an example, we demonstrate the advantages of an interactive visualization tool for multi-dimensional data. VISPLORE greatly supports the analysis of time dependent data sets, as they are produced by evolutionary optimization algorithms. We demonstrate various multi-dimensional visualization techniques, as built into VISPLORE, which help to understand the dynamics of stochastic search algorithms.},
keywords = {},
pubstate = {published},
tppubtype = {incollection}
}
von Mammen, Sebastian; Phillips, David; Davison, Timothy; Jacob, Christian
Swarm Graph Grammars: A Scale-independent, Graph-based Multi-agent Representation Conference
ANTS 2010 - 7th International Conference on Swarm Intelligence, Brussels, Belgium, 2010.
@conference{248,
title = {Swarm Graph Grammars: A Scale-independent, Graph-based Multi-agent Representation},
author = {Sebastian von Mammen and David Phillips and Timothy Davison and Christian Jacob},
year = {2010},
date = {2010-01-01},
urldate = {2010-01-01},
booktitle = {ANTS 2010 - 7th International Conference on Swarm Intelligence},
address = {Brussels, Belgium},
abstract = {<p> <p style="margin: 0.0px 0.0px 0.0px 0.0px; font: 12.0px Helvetica;">Modelling natural processes requires the implementation of an expressive representation of the involved entities and their inter- actions. We present swarm graph grammars (SGGs) as a bio-inspired modelling framework that integrates aspects of formal grammars, graph- based representation and multi-agent simulation. In SGGs, the substitution of subgraphs that represent locally defined agent interactions drive the computational process of the simulation. The generative character of formal grammars is translated into an agenttextquoterights metabolic interactions, i.e. creating or removing agents from the system. Utilizing graphs to describe interactions and relationships between pairs or sets of agents offers an easily accessible way of modelling biological phenomena. Property graphs emerge through the application of local interaction rules; we use these graphs to capture various aspects of the interaction dynamics at any given step of a simulation.</p>
</p>},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}
</p>